Parasitic worms that infect humans are not interbreeding with those that infect cattle as previously thought. This is good news for when it comes to controlling schistosomiasis, a disease caused by these worms that affects more than 200 million people globally.
For more than a decade, evidence suggested that worm species that infect humans and cattle, Schistosoma haematobium and Schistosoma bovis, were trading genes frequently – a process called hybridization. This caused concern among those working to manage schistosomiasis, because it would significantly expand the potential for infection.
But a detailed genetic analysis conducted at Texas Biomedical Research Institute (Texas Biomed) shows that – in this instance – the animal pathogen is not likely to spill over into people.
“There have been 100 or more papers talking about rampant hybridization between these different species,” said Texas Biomed Professor Tim Anderson, PhD. “This was one of the rare cases in my career where a member of my lab has written a paper that has completely changed the direction of a field.”
A long time ago
The paper, published in Nature Communications, reveals that the human parasite, Schistosoma hematobium, indeed does have some cattle parasite genes – but that this interbreeding occurred hundreds of generations ago. It is not new nor frequent.
“It’s similar to how humans have low levels of neanderthal DNA from interbreeding that occurred tens of thousands of years ago,” said Texas Biomed Staff Scientist Roy Neal Platt II, PhD. “The cattle parasite genes that remain in the human parasites likely provide some adaptive benefits to the schistosomes, but we don’t know what those benefits are yet.”
A continental divide
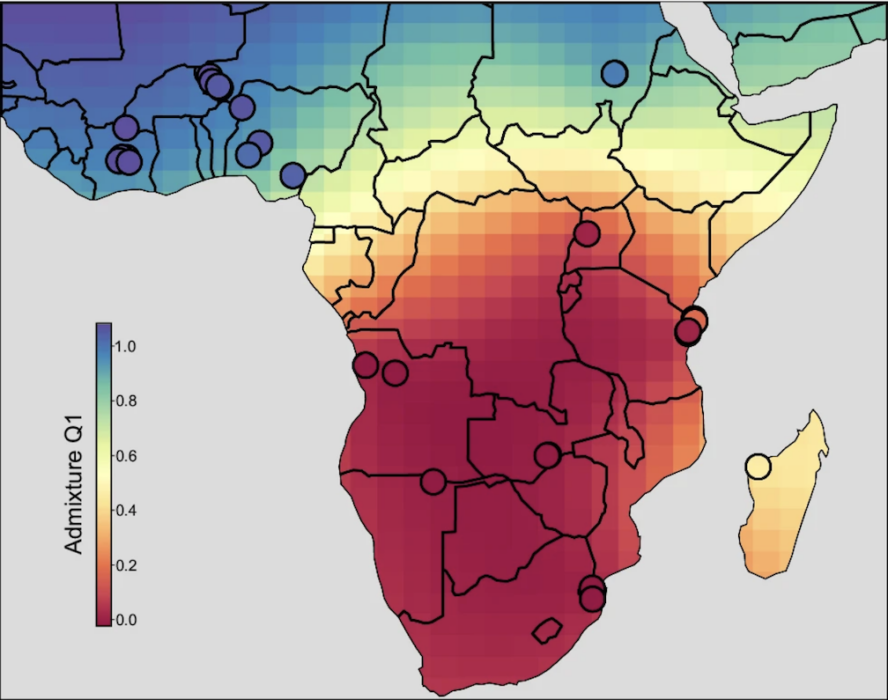
Additionally, the researchers found an unexpected divide: the human parasite samples from northern Africa contain cattle genes, but the samples from southern Africa do not.
“If you draw a line at the equator, there is a clear divide,” Dr. Platt said.
It is unclear why there are two distinct human parasite populations in Africa that do not mix. It may be due to differences in the snails found in northern versus southern Africa. Schistosome parasites have a complex life cycle that involves partially developing in snails before being released in freshwater where they then penetrate human skin to mature into
flatworms in the blood vessels. Understanding the differences between the snail hosts could help identify new ways to control the parasites.
Neglected Tropical Disease
Schistosomiasis is a Neglected Tropical Disease common in tropical and subtropical regions of the world. People are infected during routine activities – washing, swimming, fishing – in freshwater bodies infested with parasite larvae.
Once in people, the parasites mature into worms that lay eggs, which cause pain and bleeding. In children, it can cause anemia, learning problems and stunted growth. Chronic infection can lead to bloody urine, kidney failure, bladder cancer and death. The medication praziquantel is effective, though up to 30% of people are still infected after treatment.
“We hope this research will help inform how limited medical and educational resources should be targeted to be most effective as work continues to end this widespread disease,” Dr. Anderson said.
Global Collaboration

The team was able to make these findings by sequencing whole genomes – rather than just one or two genes – of 160 parasite samples from 18 countries across Africa. They evaluated more than 34 million single nucleotide variations to determine the genetic relationships between the species. The project required “decades of computation time,” which was completed in weeks at Texas Biomed’s High Performance Computing Center.
Before the first sequence could be run, there was a massive effort to gather samples of human and cow parasites from across Africa. As part of the work, Egie Enabulele, PhD, a former Texas Biomed postdoctoral scientist, traveled to collect samples from people and livestock in Nigeria. Other samples were provided by museums, including the London Natural History Museum, which consist of tiny specks of dried larvae on slips of paper. Other members of Dr. Anderson’s lab conducted extensive work to prep the samples and extract enough DNA for sequencing.
“It is easy for me to sit at the computer, run the analyses and show fancy figures, but this required so much work in the field and in the lab,” Dr. Platt stressed. “This research would not have been possible without global collaboration.”
Paper:
Platt II, R.N., Enabulele, E.E., Adeyemi, E. et al. Genomic data reveal a north-south split and introgression history of blood fluke populations across Africa. Nat Commun 16, 3508 (2025). https://doi.org/10.1038/s41467-025-58543-6
Funding: From National Institutes for Health (R01 AI166049)